Micromechanical behavior of DNA
Introduction
The topic of the project is relevant to the development of novel single-molecule manipulation techniques in biophysics and bio nanotechnology where complex DNA-liquid interactions occur. The goals of the project are to verify the proposed numerical method by com-paring the results for simple flow conditions with avail-able numerical and analytical results, to analyze the dynamics of the DNA macromolecule exposed to an uniform and shear flow, perform simulations corre-sponding to the more complex experimental flow con-ditions.
Recently the task attaching one DNA molecule spe-cifically with both ends to different microscopic con-tact pads on surfaces has been carried out for the first time with respect to the construction of DNA-templated nanowires and networks. These investigations show unambiguously that DNA molecules can be specifically integrated into microfluidic channel systems and be manipulated by hydrodynamic flows.
Because mesoscopic length scales (from nanometres to micrometers) dominate the overall behaviour of DNA, macroscopic approaches such as finite-volume or finite-element based discretizations of the continuum-flow equations, and microscopic approaches, such as molecular dynamics (MD), are not applicable since they cannot resolve the mesoscopic structures, neglect thermal fluctuation effects, or are limited to characteristic length scales near the lower bound of microfluidics. Therefore a number of research groups pursue mesoscopic approaches which can resolve DNA molecular structure, andrecover hydrodynamic forces and thermal fluctuation effects as well.
In this project we describe a use model based on smoothed dissipative particle dynamics (SDPD), which is suitable for the numerical simulation of complex flows. The method inherits the favourable properties of Smoothed Particles Hydrodynamics (SPH) [1] for complex flows and the efficient representation of mesoscopic effects. Immersed polymers are taken into account in simulations by a straight-forward modifica-tion of the SDPD-particle interactions of particles con-taining parts of the polymer. The method is validated by the comparison with theoretical results of generic cases [2].
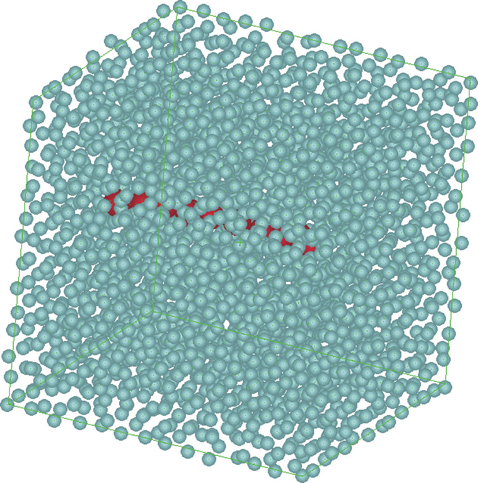
The polymer is embedded into a number of special SDPD particles (denoted as polymer beads) which represent the segments of the polymer molecule. For typical particle sizes a bead would contain mainly solvent and the volume fraction of the polymer segment is small. Therefore, polymer beads interact hydro dynamically, with additional forces due to the chemical bond between the polymer segments contained in neighbouring polymer beads. These additional forces are taken into account by a finite extensible non-linear elastic (FENE) potential
Computational details
In the influential review articles Larsen listed the fol-lowing forces important for polymer simulations. They all are captured in SDPD model:
• Viscous drag: included in SPH equations
• Entropic elasticity: FENE spring
• Brownian forces: captured by Generic formal-ism
• Hydrodynamic interaction: included via sol-vent SDPD particles.
• Excluded-volume: in SDPD particle cannot penetrate due to strong potential forces
• Internal viscosity: polymers beads are also in-teracting with normal SDPD forces
• Self-entanglement: polymer chain can en-tangle with itself but big number of beads is required.
A typical simulation procedure is illustrated in Figures 1. The polymer chain is immersed in a domain full of SDPD particles.
An implementation of the model with focus on per-formance was done in FORTRAN 90 using PPM library which hides the complexity of the parallel imple-mentation from the client author:
• PPM: Highly Efficient Parallel Particle-Mesh Library, www.mosaic.ethz.ch/Downloads/PPM
• H5Part: An API scheme to store particle data, vis.lbl.gov/Research/AcceleratorSAPP
• fgsl: FORTRAN interface to the GSL, www.lrz-muenchen.de/services/software/mathematik/gsl/fortran
The parallelization strategy is based on domain de-composition. Communication is performed using ghost-particle layer between neighbouring sub-domains. In case of PPM-client we use only a subset of mapping options provided by the library.
The performance of single processor code achieved on HLRB is around 104 particles processed per second per one processor.
Results
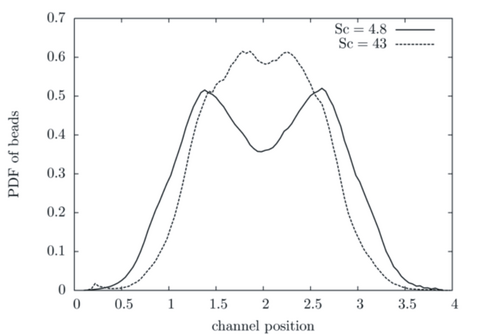
A study of polymer confined in the channel and ex-posed to a Poiseuille flow was also performed. In Fig. 2 the span-wise distribution of polymer mass is shown. We found that the profile is affected by Schmidt number: for lower Schmidt number the depletion region is pronounced at the centre of the channel; for high Schmidt number the polymer concentration tends to be higher in the centre with smaller off-centre peaks [2]. Our simulations results are generally confirmed by the recent experimental study of and more detailed mesoscale simulation.
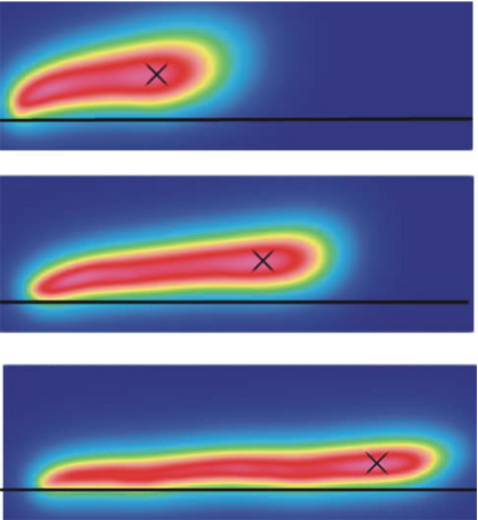
Another results are related to a DNA molecule tethered on wall surface and exposed to shear flows, it exhibits different mechanical properties due to the different hydrodynamic force and wall-chain interaction. The distributions of DNA beads in the flow-gradient plane for three cases with small, moderate and large shear rates are shown in Fig. 3.
Reference
Litvinov, S., Ellero, M., Hu, X. & Adams, N.; A splitting scheme for highly dissipative smoothed particle dynamics; Journal of Computational Physics, Elsevier, 2010, Vol. 229(15), pp. 5457-5464
Larson, R.G, The rheology of dilute solutions of flexible polymers: Progress and problems, J. Rhoelogy. 49 (2005)
Litvinov, S., Ellero, M., Hu, X. & Adams, N.; Smoothed dissipative particle dynamics model for polymer molecules in suspension; Physical Review E, APS, 2008, Vol. 77(6), pp. 66703
Litvinov, S., Hu, X.Y. and Adams, N. A. ; Numerical simulation of tethered DNA in shear flow, J. Phys.: Condens. Matter 23 (2011) 184118